LION helps Cancer researchers form hypotheses by giving them a graph based view of the research literature.
We mine Cancer abstracts from Pubmed, extract relevant biological entities from each one. Each biological entity is a node in our graph and edges between entities represent (roughly) how many abstracts the two entities appear together in.
To start with you enter a search query. Depending on the query type, you get one of three results
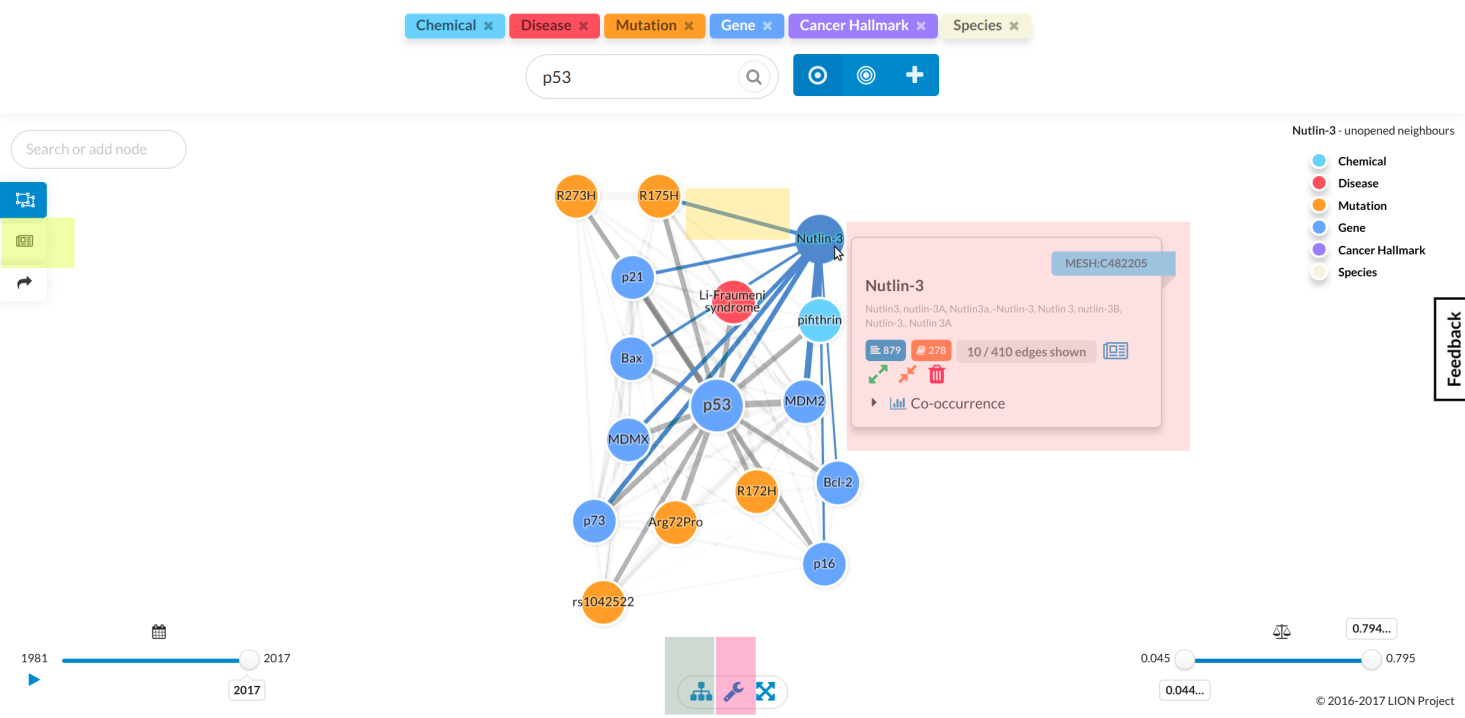
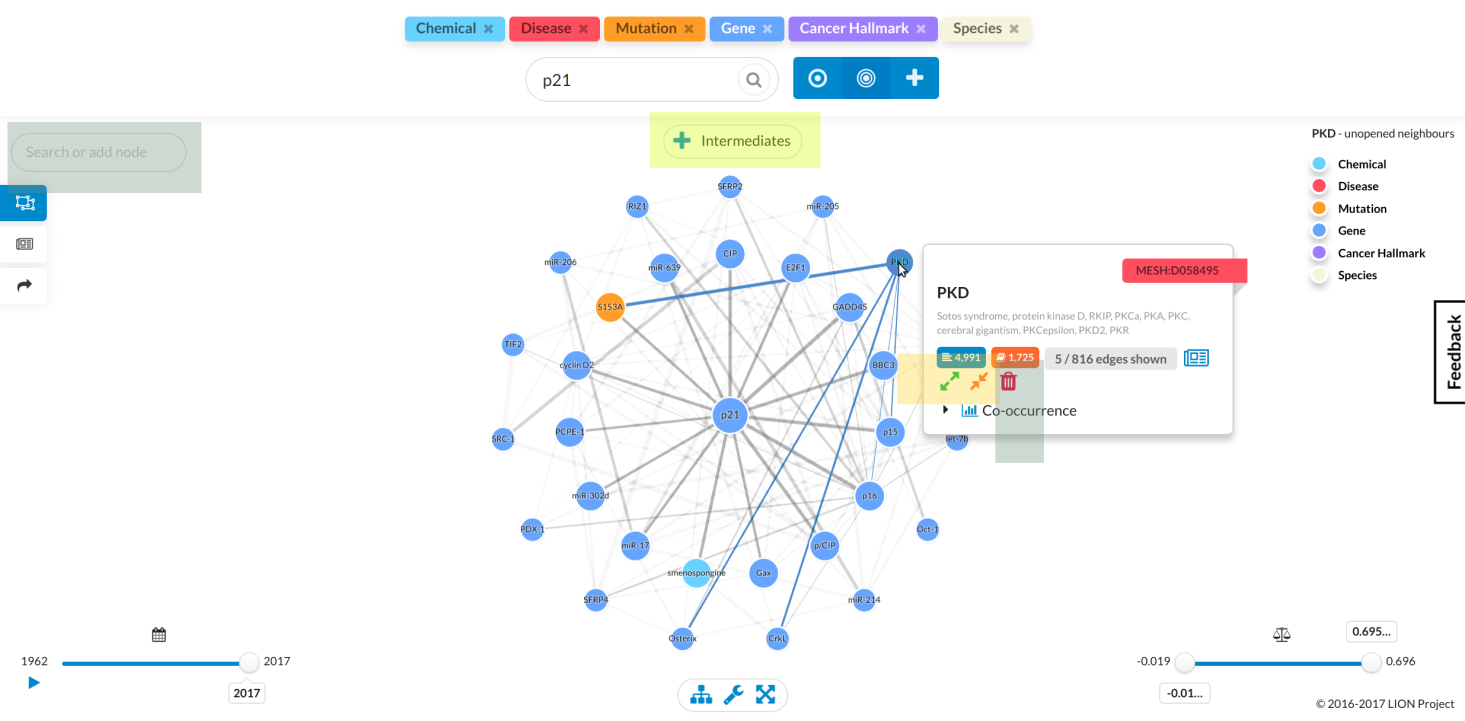
Once you've entered a query and LION returns a graph, you can start exploring it, with the following operations to help you:
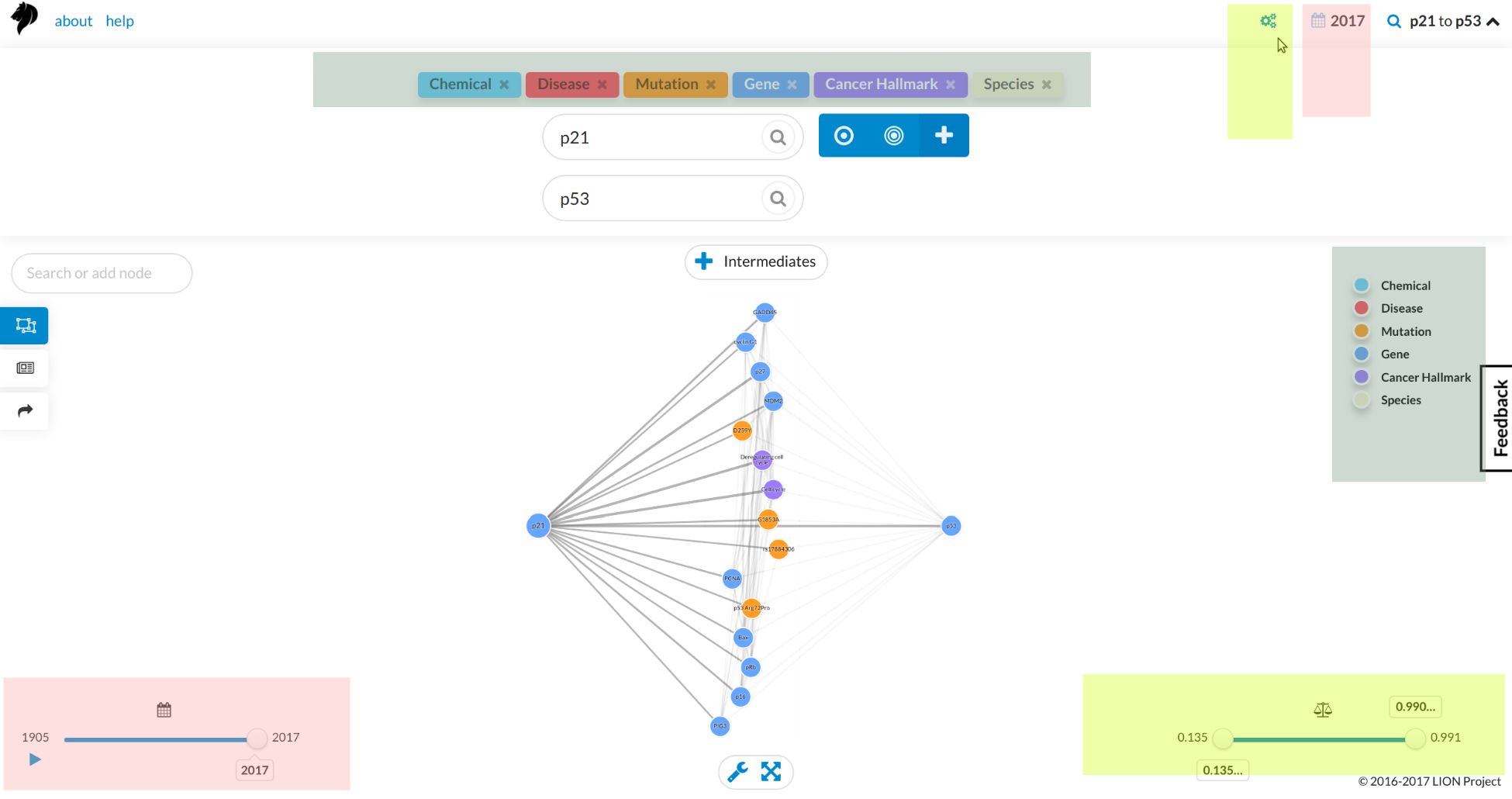
If you require further help, contact us on ltllionproject@gmail.com